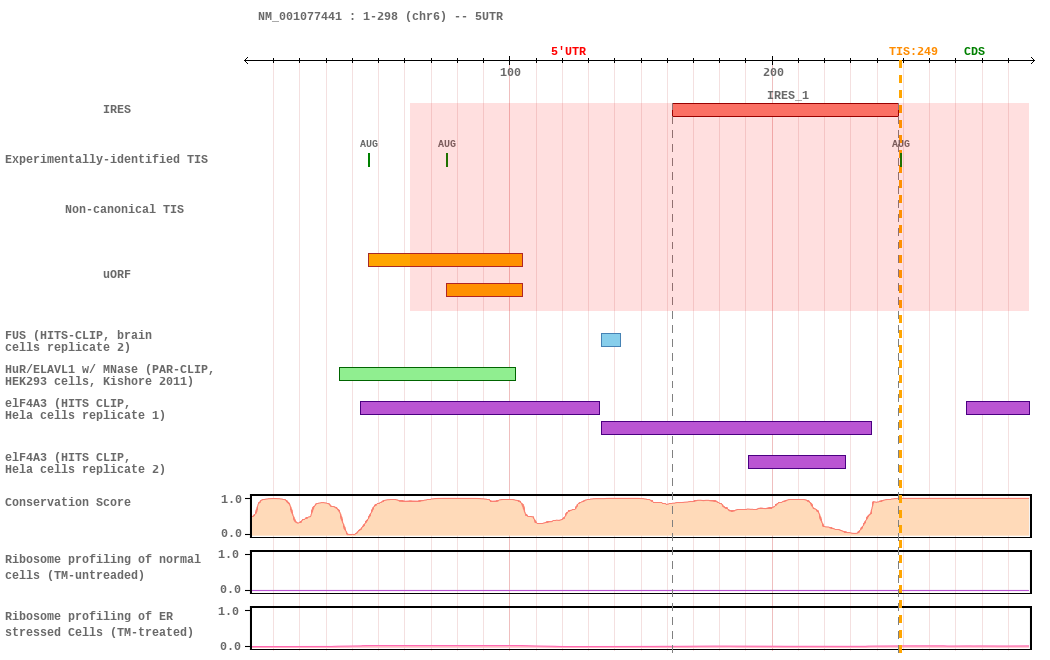
Transcript Detail IRES Map
Queried Transcript
| Transcript Info | Conservation | Conditional Translation Efficiency | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Accession | Genome Location |
Trascribed Gene |
5'UTR | CDS | Stop Codon | 3'UTR | Average Phastcon Score |
ER stressed | Normal |
ER/Normal Ratio |
| NM_001077441 | chr6: - (location) |
BCLAF1 |
248 bps (seq) |
2241 bps (seq) |
2490 ..2493 (TAA) |
4483 bps (seq) |
0.760 | 0.006 | 0.000 |
INF
(q = 0.000E+0*) |
Identified Putative IRES Elements
* indicates statistical significance:
(Thresholds
selected on a ground truth dataset)
1. Conservation Score q-value < 1.000E-6
2. RPI Score q-value < 1.000E-6.
3. Ratio (difference) q-value < 1.000E-6.
| IRES Info | Conservation |
Conditional
Translation Efficiency |
Translation Initiation-related Info Summary for the IRES | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Putative IRES | Identifying Tools |
Average Phastcon Score (ref) |
ER Stressed | Normal | ER/Normal Ratio (ref) |
# of eTIS
(ref) |
# of nTIS
(ref) |
# of uORF
(ref) |
# of ITAF Binding Events (ref) |
RPI Significance |
IRES activity (ref) |
# of Literature Evidence |
|
IRES 1 162..248 (seq) (location) |
UTRscan |
0.702
(q = 4.959E-3) |
0.007 | 0.000 |
INF
(q = 3.098E-44*) |
2 | 0 | 2 | 2 | q = 7.604E-12* | -- | 0 |
IRES-Translation Initiation Interaction Map
(TIS refers to the main ORF initiating codon)
Detailed Translation Initiation-Related Data
Phastcon Conservation Scores (Among 19 Mammalian Species)
| Interconnected IRES |
Transcript Location |
Chromosomal Location |
Conservation Score |
|---|---|---|---|
| IRES 1 (162..248) | 162 | chr6 (-): 136282660 | 0.870 |
| IRES 1 (162..248) | 163 | chr6 (-): 136282659 | 0.874 |
| IRES 1 (162..248) | 164 | chr6 (-): 136282658 | 0.884 |
| IRES 1 (162..248) | 165 | chr6 (-): 136282657 | 0.883 |
| IRES 1 (162..248) | 166 | chr6 (-): 136282656 | 0.900 |
| IRES 1 (162..248) | 167 | chr6 (-): 136282655 | 0.904 |
| IRES 1 (162..248) | 168 | chr6 (-): 136282654 | 0.909 |
| IRES 1 (162..248) | 169 | chr6 (-): 136282653 | 0.917 |
| IRES 1 (162..248) | 170 | chr6 (-): 136282652 | 0.916 |
| IRES 1 (162..248) | 171 | chr6 (-): 136282651 | 0.944 |
| IRES 1 (162..248) | 172 | chr6 (-): 136282650 | 0.952 |
| IRES 1 (162..248) | 173 | chr6 (-): 136282649 | 0.953 |
| IRES 1 (162..248) | 174 | chr6 (-): 136282648 | 0.946 |
| IRES 1 (162..248) | 175 | chr6 (-): 136282647 | 0.952 |
| IRES 1 (162..248) | 176 | chr6 (-): 136282646 | 0.951 |
| IRES 1 (162..248) | 177 | chr6 (-): 136282645 | 0.943 |
| IRES 1 (162..248) | 178 | chr6 (-): 136282644 | 0.937 |
| IRES 1 (162..248) | 179 | chr6 (-): 136282643 | 0.923 |
| IRES 1 (162..248) | 180 | chr6 (-): 136282642 | 0.879 |
| IRES 1 (162..248) | 181 | chr6 (-): 136282641 | 0.841 |
| IRES 1 (162..248) | 182 | chr6 (-): 136282640 | 0.753 |
| IRES 1 (162..248) | 183 | chr6 (-): 136282639 | 0.733 |
| IRES 1 (162..248) | 184 | chr6 (-): 136282638 | 0.668 |
| IRES 1 (162..248) | 185 | chr6 (-): 136282637 | 0.654 |
| IRES 1 (162..248) | 186 | chr6 (-): 136282636 | 0.670 |
| IRES 1 (162..248) | 187 | chr6 (-): 136282635 | 0.690 |
| IRES 1 (162..248) | 188 | chr6 (-): 136282634 | 0.691 |
| IRES 1 (162..248) | 189 | chr6 (-): 136282633 | 0.696 |
| IRES 1 (162..248) | 190 | chr6 (-): 136282632 | 0.702 |
| IRES 1 (162..248) | 191 | chr6 (-): 136282631 | 0.712 |
| IRES 1 (162..248) | 192 | chr6 (-): 136282630 | 0.707 |
| IRES 1 (162..248) | 193 | chr6 (-): 136282629 | 0.707 |
| IRES 1 (162..248) | 194 | chr6 (-): 136282628 | 0.689 |
| IRES 1 (162..248) | 195 | chr6 (-): 136282627 | 0.721 |
| IRES 1 (162..248) | 196 | chr6 (-): 136282626 | 0.728 |
| IRES 1 (162..248) | 197 | chr6 (-): 136282625 | 0.721 |
| IRES 1 (162..248) | 198 | chr6 (-): 136282624 | 0.718 |
| IRES 1 (162..248) | 199 | chr6 (-): 136282623 | 0.736 |
| IRES 1 (162..248) | 200 | chr6 (-): 136282622 | 0.737 |
| IRES 1 (162..248) | 201 | chr6 (-): 136282621 | 0.766 |
| IRES 1 (162..248) | 202 | chr6 (-): 136282620 | 0.836 |
| IRES 1 (162..248) | 203 | chr6 (-): 136282619 | 0.889 |
| IRES 1 (162..248) | 204 | chr6 (-): 136282618 | 0.906 |
| IRES 1 (162..248) | 205 | chr6 (-): 136282617 | 0.928 |
| IRES 1 (162..248) | 206 | chr6 (-): 136282616 | 0.957 |
| IRES 1 (162..248) | 207 | chr6 (-): 136282615 | 0.966 |
| IRES 1 (162..248) | 208 | chr6 (-): 136282614 | 0.968 |
| IRES 1 (162..248) | 209 | chr6 (-): 136282613 | 0.971 |
| IRES 1 (162..248) | 210 | chr6 (-): 136282612 | 0.976 |
| IRES 1 (162..248) | 211 | chr6 (-): 136282611 | 0.976 |
| IRES 1 (162..248) | 212 | chr6 (-): 136282610 | 0.971 |
| IRES 1 (162..248) | 213 | chr6 (-): 136282609 | 0.960 |
| IRES 1 (162..248) | 214 | chr6 (-): 136282608 | 0.935 |
| IRES 1 (162..248) | 215 | chr6 (-): 136282607 | 0.875 |
| IRES 1 (162..248) | 216 | chr6 (-): 136282606 | 0.730 |
| IRES 1 (162..248) | 217 | chr6 (-): 136282605 | 0.704 |
| IRES 1 (162..248) | 218 | chr6 (-): 136282604 | 0.627 |
| IRES 1 (162..248) | 219 | chr6 (-): 136282603 | 0.395 |
| IRES 1 (162..248) | 220 | chr6 (-): 136282602 | 0.216 |
| IRES 1 (162..248) | 221 | chr6 (-): 136282601 | 0.208 |
| IRES 1 (162..248) | 222 | chr6 (-): 136282600 | 0.204 |
| IRES 1 (162..248) | 223 | chr6 (-): 136282599 | 0.178 |
| IRES 1 (162..248) | 224 | chr6 (-): 136282598 | 0.157 |
| IRES 1 (162..248) | 225 | chr6 (-): 136282597 | 0.141 |
| IRES 1 (162..248) | 226 | chr6 (-): 136282596 | 0.129 |
| IRES 1 (162..248) | 227 | chr6 (-): 136282595 | 0.095 |
| IRES 1 (162..248) | 228 | chr6 (-): 136282594 | 0.070 |
| IRES 1 (162..248) | 229 | chr6 (-): 136282593 | 0.050 |
| IRES 1 (162..248) | 230 | chr6 (-): 136282592 | 0.049 |
| IRES 1 (162..248) | 231 | chr6 (-): 136282591 | 0.037 |
| IRES 1 (162..248) | 232 | chr6 (-): 136282590 | 0.036 |
| IRES 1 (162..248) | 233 | chr6 (-): 136282589 | 0.041 |
| IRES 1 (162..248) | 234 | chr6 (-): 136282588 | 0.131 |
| IRES 1 (162..248) | 235 | chr6 (-): 136282587 | 0.240 |
| IRES 1 (162..248) | 236 | chr6 (-): 136282586 | 0.371 |
| IRES 1 (162..248) | 237 | chr6 (-): 136282585 | 0.522 |
| IRES 1 (162..248) | 238 | chr6 (-): 136282584 | 0.573 |
| IRES 1 (162..248) | 239 | chr6 (-): 136279876 | 0.915 |
| IRES 1 (162..248) | 240 | chr6 (-): 136279875 | 0.902 |
| IRES 1 (162..248) | 241 | chr6 (-): 136279874 | 0.913 |
| IRES 1 (162..248) | 242 | chr6 (-): 136279873 | 0.932 |
| IRES 1 (162..248) | 243 | chr6 (-): 136279872 | 0.956 |
| IRES 1 (162..248) | 244 | chr6 (-): 136279871 | 0.964 |
| IRES 1 (162..248) | 245 | chr6 (-): 136279870 | 0.975 |
| IRES 1 (162..248) | 246 | chr6 (-): 136279869 | 0.989 |
| IRES 1 (162..248) | 247 | chr6 (-): 136279868 | 0.996 |
| IRES 1 (162..248) | 248 | chr6 (-): 136279867 | 0.998 |
Potential IRES Structural Form
IRES 1
(162.. 248)
Best Predicted IRES Structural Form:
.........(((((.((......((((((((.(((...((((..((....))..))))))).)))..)))))))..)))))......
Predicted by: SPARSE
Structural RPI Interpretability Significance: q = 7.604E-12*
(Visualization)
Conditional Translation Activity
ER-Stressed Ribisome Profiling (TM-treated)
| Interconnected IRES |
Transcript Location |
Chromosomal Location |
Stressed Translation Activity |
|---|---|---|---|
| IRES 1 (162..248) | 162 | chr6 (-): 136282660 | 0.00472548425619 |
| IRES 1 (162..248) | 163 | chr6 (-): 136282659 | 0.0048779225165 |
| IRES 1 (162..248) | 164 | chr6 (-): 136282658 | 0.0048779225165 |
| IRES 1 (162..248) | 165 | chr6 (-): 136282657 | 0.0048779225165 |
| IRES 1 (162..248) | 166 | chr6 (-): 136282656 | 0.00533523729744 |
| IRES 1 (162..248) | 167 | chr6 (-): 136282655 | 0.00518279903713 |
| IRES 1 (162..248) | 168 | chr6 (-): 136282654 | 0.00579253641476 |
| IRES 1 (162..248) | 169 | chr6 (-): 136282653 | 0.00609740510358 |
| IRES 1 (162..248) | 170 | chr6 (-): 136282652 | 0.00701202683364 |
| IRES 1 (162..248) | 171 | chr6 (-): 136282651 | 0.00762177987489 |
| IRES 1 (162..248) | 172 | chr6 (-): 136282650 | 0.00853635461412 |
| IRES 1 (162..248) | 173 | chr6 (-): 136282649 | 0.00868883986527 |
| IRES 1 (162..248) | 174 | chr6 (-): 136282648 | 0.00884124679836 |
| IRES 1 (162..248) | 175 | chr6 (-): 136282647 | 0.00899365373145 |
| IRES 1 (162..248) | 176 | chr6 (-): 136282646 | 0.00899365373145 |
| IRES 1 (162..248) | 177 | chr6 (-): 136282645 | 0.00929854591568 |
| IRES 1 (162..248) | 178 | chr6 (-): 136282644 | 0.00929854591568 |
| IRES 1 (162..248) | 179 | chr6 (-): 136282643 | 0.00929854591568 |
| IRES 1 (162..248) | 180 | chr6 (-): 136282642 | 0.00945095284877 |
| IRES 1 (162..248) | 181 | chr6 (-): 136282641 | 0.00960343809992 |
| IRES 1 (162..248) | 182 | chr6 (-): 136282640 | 0.0100607372172 |
| IRES 1 (162..248) | 183 | chr6 (-): 136282639 | 0.0105180363346 |
| IRES 1 (162..248) | 184 | chr6 (-): 136282638 | 0.0105180363346 |
| IRES 1 (162..248) | 185 | chr6 (-): 136282637 | 0.00975584503301 |
| IRES 1 (162..248) | 186 | chr6 (-): 136282636 | 0.00884124679836 |
| IRES 1 (162..248) | 187 | chr6 (-): 136282635 | 0.0080790554968 |
| IRES 1 (162..248) | 188 | chr6 (-): 136282634 | 0.00792664856371 |
| IRES 1 (162..248) | 189 | chr6 (-): 136282633 | 0.00746934944639 |
| IRES 1 (162..248) | 190 | chr6 (-): 136282632 | 0.00746934944639 |
| IRES 1 (162..248) | 191 | chr6 (-): 136282631 | 0.0076217877067 |
| IRES 1 (162..248) | 192 | chr6 (-): 136282630 | 0.00746934944639 |
| IRES 1 (162..248) | 193 | chr6 (-): 136282629 | 0.00746934944639 |
| IRES 1 (162..248) | 194 | chr6 (-): 136282628 | 0.00746934944639 |
| IRES 1 (162..248) | 195 | chr6 (-): 136282627 | 0.00731691118607 |
| IRES 1 (162..248) | 196 | chr6 (-): 136282626 | 0.00716448075757 |
| IRES 1 (162..248) | 197 | chr6 (-): 136282625 | 0.00716448075757 |
| IRES 1 (162..248) | 198 | chr6 (-): 136282624 | 0.00701204249726 |
| IRES 1 (162..248) | 199 | chr6 (-): 136282623 | 0.00670717380844 |
| IRES 1 (162..248) | 200 | chr6 (-): 136282622 | 0.00655474337993 |
| IRES 1 (162..248) | 201 | chr6 (-): 136282621 | 0.00670718164024 |
| IRES 1 (162..248) | 202 | chr6 (-): 136282620 | 0.00670718164024 |
| IRES 1 (162..248) | 203 | chr6 (-): 136282619 | 0.006097428599 |
| IRES 1 (162..248) | 204 | chr6 (-): 136282618 | 0.00487793818011 |
| IRES 1 (162..248) | 205 | chr6 (-): 136282617 | 0.00503036860862 |
| IRES 1 (162..248) | 206 | chr6 (-): 136282616 | 0.0047254999198 |
| IRES 1 (162..248) | 207 | chr6 (-): 136282615 | 0.00426819297067 |
| IRES 1 (162..248) | 208 | chr6 (-): 136282614 | 0.00426819297067 |
| IRES 1 (162..248) | 209 | chr6 (-): 136282613 | 0.00442062339918 |
| IRES 1 (162..248) | 210 | chr6 (-): 136282612 | 0.004725492088 |
| IRES 1 (162..248) | 211 | chr6 (-): 136282611 | 0.00518280686893 |
| IRES 1 (162..248) | 212 | chr6 (-): 136282610 | 0.00564010598626 |
| IRES 1 (162..248) | 213 | chr6 (-): 136282609 | 0.00548767555775 |
| IRES 1 (162..248) | 214 | chr6 (-): 136282608 | 0.00533524512924 |
| IRES 1 (162..248) | 215 | chr6 (-): 136282607 | 0.00518279903713 |
| IRES 1 (162..248) | 216 | chr6 (-): 136282606 | 0.0048779225165 |
| IRES 1 (162..248) | 217 | chr6 (-): 136282605 | 0.00396330861824 |
| IRES 1 (162..248) | 218 | chr6 (-): 136282604 | 0.00381087818974 |
| IRES 1 (162..248) | 219 | chr6 (-): 136282603 | 0.00381087818974 |
| IRES 1 (162..248) | 220 | chr6 (-): 136282602 | 0.00365843992943 |
| IRES 1 (162..248) | 221 | chr6 (-): 136282601 | 0.00365843992943 |
| IRES 1 (162..248) | 222 | chr6 (-): 136282600 | 0.00365843992943 |
| IRES 1 (162..248) | 223 | chr6 (-): 136282599 | 0.00365843992943 |
| IRES 1 (162..248) | 224 | chr6 (-): 136282598 | 0.00365843992943 |
| IRES 1 (162..248) | 225 | chr6 (-): 136282597 | 0.00396330861824 |
| IRES 1 (162..248) | 226 | chr6 (-): 136282596 | 0.00411573904675 |
| IRES 1 (162..248) | 227 | chr6 (-): 136282595 | 0.00396331645005 |
| IRES 1 (162..248) | 228 | chr6 (-): 136282594 | 0.00426818513887 |
| IRES 1 (162..248) | 229 | chr6 (-): 136282593 | 0.00442061556737 |
| IRES 1 (162..248) | 230 | chr6 (-): 136282592 | 0.0048779146847 |
| IRES 1 (162..248) | 231 | chr6 (-): 136282591 | 0.00503035294501 |
| IRES 1 (162..248) | 232 | chr6 (-): 136282590 | 0.00503035294501 |
| IRES 1 (162..248) | 233 | chr6 (-): 136282589 | 0.0048779146847 |
| IRES 1 (162..248) | 234 | chr6 (-): 136282588 | 0.00487790685289 |
| IRES 1 (162..248) | 235 | chr6 (-): 136282587 | 0.00533521380202 |
| IRES 1 (162..248) | 236 | chr6 (-): 136282586 | 0.0048779225165 |
| IRES 1 (162..248) | 237 | chr6 (-): 136282585 | 0.00853635461412 |
| IRES 1 (162..248) | 238 | chr6 (-): 136282584 | 0.00929846759763 |
| IRES 1 (162..248) | 239 | chr6 (-): 136279876 | 0.0099082519661 |
| IRES 1 (162..248) | 240 | chr6 (-): 136279875 | 0.0099082519661 |
| IRES 1 (162..248) | 241 | chr6 (-): 136279874 | 0.0099082519661 |
| IRES 1 (162..248) | 242 | chr6 (-): 136279873 | 0.0099082519661 |
| IRES 1 (162..248) | 243 | chr6 (-): 136279872 | 0.00945095284877 |
| IRES 1 (162..248) | 244 | chr6 (-): 136279871 | 0.0100606588992 |
| IRES 1 (162..248) | 245 | chr6 (-): 136279870 | 0.011127742385 |
| IRES 1 (162..248) | 246 | chr6 (-): 136279869 | 0.0115850415023 |
| IRES 1 (162..248) | 247 | chr6 (-): 136279868 | 0.0114326345692 |
| IRES 1 (162..248) | 248 | chr6 (-): 136279867 | 0.0115850415023 |
Normal Ribisome Profiling (TM-untreaed)
| Interconnected IRES |
Transcript Location |
Chromosomal Location |
Normal Translation Activity |
|---|---|---|---|
| IRES 1 (162..248) | 162 | chr6 (-): 136282660 | 0 |
| IRES 1 (162..248) | 163 | chr6 (-): 136282659 | 0 |
| IRES 1 (162..248) | 164 | chr6 (-): 136282658 | 0 |
| IRES 1 (162..248) | 165 | chr6 (-): 136282657 | 0 |
| IRES 1 (162..248) | 166 | chr6 (-): 136282656 | 0 |
| IRES 1 (162..248) | 167 | chr6 (-): 136282655 | 0 |
| IRES 1 (162..248) | 168 | chr6 (-): 136282654 | 0 |
| IRES 1 (162..248) | 169 | chr6 (-): 136282653 | 0 |
| IRES 1 (162..248) | 170 | chr6 (-): 136282652 | 0 |
| IRES 1 (162..248) | 171 | chr6 (-): 136282651 | 0 |
| IRES 1 (162..248) | 172 | chr6 (-): 136282650 | 0 |
| IRES 1 (162..248) | 173 | chr6 (-): 136282649 | 0 |
| IRES 1 (162..248) | 174 | chr6 (-): 136282648 | 0 |
| IRES 1 (162..248) | 175 | chr6 (-): 136282647 | 0 |
| IRES 1 (162..248) | 176 | chr6 (-): 136282646 | 0 |
| IRES 1 (162..248) | 177 | chr6 (-): 136282645 | 0 |
| IRES 1 (162..248) | 178 | chr6 (-): 136282644 | 0 |
| IRES 1 (162..248) | 179 | chr6 (-): 136282643 | 0 |
| IRES 1 (162..248) | 180 | chr6 (-): 136282642 | 0 |
| IRES 1 (162..248) | 181 | chr6 (-): 136282641 | 0 |
| IRES 1 (162..248) | 182 | chr6 (-): 136282640 | 0 |
| IRES 1 (162..248) | 183 | chr6 (-): 136282639 | 0 |
| IRES 1 (162..248) | 184 | chr6 (-): 136282638 | 0 |
| IRES 1 (162..248) | 185 | chr6 (-): 136282637 | 0 |
| IRES 1 (162..248) | 186 | chr6 (-): 136282636 | 0 |
| IRES 1 (162..248) | 187 | chr6 (-): 136282635 | 0 |
| IRES 1 (162..248) | 188 | chr6 (-): 136282634 | 0 |
| IRES 1 (162..248) | 189 | chr6 (-): 136282633 | 0 |
| IRES 1 (162..248) | 190 | chr6 (-): 136282632 | 0 |
| IRES 1 (162..248) | 191 | chr6 (-): 136282631 | 0 |
| IRES 1 (162..248) | 192 | chr6 (-): 136282630 | 0 |
| IRES 1 (162..248) | 193 | chr6 (-): 136282629 | 0 |
| IRES 1 (162..248) | 194 | chr6 (-): 136282628 | 0 |
| IRES 1 (162..248) | 195 | chr6 (-): 136282627 | 0 |
| IRES 1 (162..248) | 196 | chr6 (-): 136282626 | 0 |
| IRES 1 (162..248) | 197 | chr6 (-): 136282625 | 0 |
| IRES 1 (162..248) | 198 | chr6 (-): 136282624 | 0 |
| IRES 1 (162..248) | 199 | chr6 (-): 136282623 | 0 |
| IRES 1 (162..248) | 200 | chr6 (-): 136282622 | 0 |
| IRES 1 (162..248) | 201 | chr6 (-): 136282621 | 0 |
| IRES 1 (162..248) | 202 | chr6 (-): 136282620 | 0 |
| IRES 1 (162..248) | 203 | chr6 (-): 136282619 | 0 |
| IRES 1 (162..248) | 204 | chr6 (-): 136282618 | 0 |
| IRES 1 (162..248) | 205 | chr6 (-): 136282617 | 0 |
| IRES 1 (162..248) | 206 | chr6 (-): 136282616 | 0 |
| IRES 1 (162..248) | 207 | chr6 (-): 136282615 | 0 |
| IRES 1 (162..248) | 208 | chr6 (-): 136282614 | 0 |
| IRES 1 (162..248) | 209 | chr6 (-): 136282613 | 0 |
| IRES 1 (162..248) | 210 | chr6 (-): 136282612 | 0 |
| IRES 1 (162..248) | 211 | chr6 (-): 136282611 | 0 |
| IRES 1 (162..248) | 212 | chr6 (-): 136282610 | 0 |
| IRES 1 (162..248) | 213 | chr6 (-): 136282609 | 0 |
| IRES 1 (162..248) | 214 | chr6 (-): 136282608 | 0 |
| IRES 1 (162..248) | 215 | chr6 (-): 136282607 | 0 |
| IRES 1 (162..248) | 216 | chr6 (-): 136282606 | 0 |
| IRES 1 (162..248) | 217 | chr6 (-): 136282605 | 0 |
| IRES 1 (162..248) | 218 | chr6 (-): 136282604 | 0 |
| IRES 1 (162..248) | 219 | chr6 (-): 136282603 | 0 |
| IRES 1 (162..248) | 220 | chr6 (-): 136282602 | 0 |
| IRES 1 (162..248) | 221 | chr6 (-): 136282601 | 0 |
| IRES 1 (162..248) | 222 | chr6 (-): 136282600 | 0 |
| IRES 1 (162..248) | 223 | chr6 (-): 136282599 | 0 |
| IRES 1 (162..248) | 224 | chr6 (-): 136282598 | 0 |
| IRES 1 (162..248) | 225 | chr6 (-): 136282597 | 0 |
| IRES 1 (162..248) | 226 | chr6 (-): 136282596 | 0 |
| IRES 1 (162..248) | 227 | chr6 (-): 136282595 | 0 |
| IRES 1 (162..248) | 228 | chr6 (-): 136282594 | 0 |
| IRES 1 (162..248) | 229 | chr6 (-): 136282593 | 0 |
| IRES 1 (162..248) | 230 | chr6 (-): 136282592 | 0 |
| IRES 1 (162..248) | 231 | chr6 (-): 136282591 | 0 |
| IRES 1 (162..248) | 232 | chr6 (-): 136282590 | 0 |
| IRES 1 (162..248) | 233 | chr6 (-): 136282589 | 0 |
| IRES 1 (162..248) | 234 | chr6 (-): 136282588 | 0 |
| IRES 1 (162..248) | 235 | chr6 (-): 136282587 | 0 |
| IRES 1 (162..248) | 236 | chr6 (-): 136282586 | 0 |
| IRES 1 (162..248) | 237 | chr6 (-): 136282585 | 0 |
| IRES 1 (162..248) | 238 | chr6 (-): 136282584 | 0 |
| IRES 1 (162..248) | 239 | chr6 (-): 136279876 | 0 |
| IRES 1 (162..248) | 240 | chr6 (-): 136279875 | 0 |
| IRES 1 (162..248) | 241 | chr6 (-): 136279874 | 0 |
| IRES 1 (162..248) | 242 | chr6 (-): 136279873 | 0 |
| IRES 1 (162..248) | 243 | chr6 (-): 136279872 | 0 |
| IRES 1 (162..248) | 244 | chr6 (-): 136279871 | 0 |
| IRES 1 (162..248) | 245 | chr6 (-): 136279870 | 0 |
| IRES 1 (162..248) | 246 | chr6 (-): 136279869 | 0 |
| IRES 1 (162..248) | 247 | chr6 (-): 136279868 | 0 |
| IRES 1 (162..248) | 248 | chr6 (-): 136279867 | 0 |
Experimentally-Probed TIS (eTIS)
| Interconnected IRES | TIS Transcript Location |
TIS Chromosomal Location |
Codon |
|---|---|---|---|
| IRES 1 (162..248) | 76..78 |
chr6 (-) : 136289769 - 136289771. |
AUG |
| IRES 1 (162..248) | 249..251 |
chr6 (-) : 136279864 - 136279866. |
AUG |
Predicted Non-canonical TIS (nTIS)
| Interconnected IRES | TIS Transcript Location |
TIS Chromosomal Location |
Non-canonical Start Codon |
Confidence |
|---|
uORF
| Interconnected IRES |
uORF Transcript Location |
uORF Chromosomal Location |
uORF Sequence | Sequence |
|---|---|---|---|---|
| IRES 1 (162..248) | 46..105 |
chr6 (-) : 136289742 - 136289801. |
60 bps (seq) | AUGGUUUUCCUCGCGUUCUUGAGUCGGGAAAUGGCCGCUGUGUGGUUGCAACGGAGAUAA |
| IRES 1 (162..248) | 76..105 |
chr6 (-) : 136289742 - 136289771. |
30 bps (seq) | AUGGCCGCUGUGUGGUUGCAACGGAGAUAA |
ITAF Binding Events
| Interconnected IRES |
ITAF Type | ITAF Transcript Location |
ITAF Chromosomal Location |
ITAF Sequence |
Sequence | Identified Score |
|---|---|---|---|---|---|---|
| IRES 1 (162..248) | eIF4A3 (HITS CLIP, Hela cells replicate 1) | 135..238 |
chr6 (-) : 136282584 - 136282687. |
104 bps (seq) | GAAUUCGAAUUUAGAGUUUAAUUUCUCAGAGCAUUCUCUCCAGGAAGAAUUUUUACAGUAUCUCAAAGACUUCACUUGACUUCUUGAUCCUGCAUAAAACCAAG | 14 |
| IRES 1 (162..248) | eIF4A3 (HITS CLIP, Hela cells replicate 2) | 191..228 |
chr6 (-) : 136282594 - 136282631. |
38 bps (seq) | AGUAUCUCAAAGACUUCACUUGACUUCUUGAUCCUGCA | 3 |
IRES Activity Measurements
| Interconnected IRES |
IRES Activity |
Promoter Activity |
Splicing Activity |
Matched Transcript Location |
Matched Transcript Sequence | Matched Chromosomal Location |
Designed Human Oligo Sequence |
Designed Oligo Sequence | BLASTN Query Cover |
BLASTN Percent Identity |
|---|
IRES Elements Verified in the Literature
| Interconnected IRES |
Matched Transcript Location |
Matched Transcript Sequence | Matched Chromosomal Location |
Literature Verified Sequence |
Verified Sequence | BLASTN Query Cover |
BLASTN Percent Identity |
Reference |
|---|
Download
The track data of this page can be download via the following two formats:
- Copyright © 2020, NUK All rights reserved. Computational Biology & Intelligence System Lab